Install & Guide
𝘚𝘶𝘱𝘱𝘰𝘳𝘵 𝘵𝘩𝘦 𝘰𝘱𝘦𝘯 𝘴𝘰𝘶𝘳𝘤𝘦 𝘮𝘰𝘷𝘦𝘮𝘦𝘯𝘵 𝘵𝘰 𝘢𝘭𝘭𝘦𝘷𝘪𝘢𝘵𝘦 𝘸𝘰𝘳𝘬 𝘱𝘳𝘦𝘴𝘴𝘶𝘳𝘦 𝘧𝘰𝘳 𝘴𝘤𝘪𝘦𝘯𝘵𝘪𝘧𝘪𝘤 𝘱𝘦𝘦𝘳𝘴, 𝘦𝘭𝘪𝘮𝘪𝘯𝘢𝘵𝘦 𝘵𝘦𝘤𝘩𝘯𝘪𝘤𝘢𝘭 𝘣𝘢𝘳𝘳𝘪𝘦𝘳𝘴, 𝘢𝘯𝘥 𝘱𝘳𝘰𝘮𝘰𝘵𝘦 𝘪𝘯𝘵𝘦𝘳𝘥𝘪𝘴𝘤𝘪𝘱𝘭𝘪𝘯𝘢𝘳𝘺 𝘤𝘰𝘭𝘭𝘢𝘣𝘰𝘳𝘢𝘵𝘪𝘰𝘯.
Note: To protect the originality of the manuscript, we will release the password for the key functional package during peer review, public person will have it after peer review has been completed.
1.Dependencies
Python = 3.9.18(No compatibility testing performed)
Matlab = R2024a(In theory, all versions are supported, because Matlab is backward compatible)
—–Matlab Tool Box = Parllel computing toolbox
Statics and machine learning toolbox
R = 4.3.2
Anaconda/Miniconda (Highly recommend)
2.PIP Install
pip install LittleSnowFox
3.Download the database
After installing LittleSnowFox using pip install LittleSnowFox, the Matlab program will be successfully downloaded. However, the package does not include the accompanying data files. To avoid potential errors in downloading data files through the Python component, you need to manually download the data files. You can directly download the entire database folder.
If the folder is placed correctly, you will be prompted with “Do you want to overwrite the database folder?” Select “Yes” to write the data files. Ensure that the data directory is correctly overwritten in the Anaconda environment during the process.
#Database folder Structure:
---[LittleSnowFox]
---[kl_sample]
---[workcatalogue]
---[database]
---[Clustering_sample]
---[e-cigarettes], [e-cigarettes_difference_expression], [fibroblasts]
---[data] #File processed by Jupyter Notebook
---[Rdata] #File processed by R
---[result] #File processed by Matlab
---main_v3_[Sample Name].m #Matlab Main Coding
---[Tracing_sample]
---[Hematopoiesis], [Nerveferroptosis], [Reprogramming]
---[data] #File processed by Jupyter Notebook
---[Rdata] #File processed by R
---[result] #File processed by Matlab
---[R_processing] #Preprocessing with R
---Hematopoiesis_all.R
---Hematopoiesis_prog.R
---R-Mature-Myocardial.R
---R-Mature-MI Tissue.R
---R-Mature-Ferroptosis.R
---Reprogramming_all.R
---Reprogramming_prog.R
---cigarette.ipynb #First step for e-cigarette analysis
---fibroblasts.ipynb #First step for fibroblasts analysis
---neuron.ipynb #First step for neuron analysis
---tracing.ipynb #First step for tracing analysis
3.1.Database & Matlab processing codes
https://drive.google.com/drive/folders/1tt8eXFgE1P7giZ94zG37vByWgZpvUtkN?usp=drive_link
When installing TOGGLE (KALIN) using Anaconda, the path is typically:
./anaconda3/envs/[Your Environment Name]/Lib/site-packages/LittleSnowFox
Overwrite the files in
./LittleSnowFox/
3.2.R code
#link
https://drive.google.com/drive/folders/1tt8eXFgE1P7giZ94zG37vByWgZpvUtkN?usp=drive_link
3.2.R package
#link
https://drive.google.com/drive/folders/15NtoVXZbAn05JSMTRWEnWAn6eyNqXrng?usp=drive_link
We provide GEO dataset backup and R language library file backup
4.Cover Letter
𝑩𝒊𝒐𝒍𝒐𝒈𝒚 𝑪𝒉𝒂𝒍𝒍𝒆𝒏𝒈𝒆𝒔:
Terminally differentiated cells undergo programmed cell death through distinct molecular pathways, including ferroptosis, apoptosis, and pyroptosis. However, identifying specific types remains challenging due to the complexity of signaling pathways, marker genes, and the limitations of current tools.
𝑨𝒍𝒈𝒐𝒓𝒊𝒕𝒉𝒎 𝑪𝒉𝒂𝒍𝒍𝒆𝒏𝒈𝒆𝒔:
It is necessary to use unsupervised algorithms because the function and lineage changes of cells are a continuous process. If it is possible to obtain a broadly representative set of labeled samples that cover the entire fate map, then the status of cells on the fate map is known, eliminating the need for differentiation between function and lineage. The current cell tracking markers are not only expensive but also cannot be used on mature cells that no longer divide.
Even when addressing the same problem repeatedly, it is difficult to combine single-cell RNA transcription maps due to the inherent systematic bias characteristics of biological experiments, which often make supervised training infeasible.
which presents the following issues:
• Endpoint loss (e.g., missing terminal samples like dead neurons in stroke).
• Time loss (difficulty in sampling at multiple intervals).
• Sample imbalance (uneven representation of cell stages), and the absence of clear fate boundaries (gradual transitions in processes like ferroptosis).
• Lack of Boundaries: cell soft and same type and subtype lack type boundaries, fate prediction of neurons lacks clear boundaries between cell states
𝑰𝒏𝒏𝒐𝒗𝒂𝒕𝒊𝒐𝒏 𝒐𝒇 𝑴𝒆𝒕𝒉𝒐𝒅𝒔:
- Based on the ability to perform mature cell subtype/type classification and lineage tracing prediction, we extended functionality to enable classification within the same subtype/type of cells based on their specific functional states.
- While existing techniques can only conduct subtype classification and lineage tracing, we enhanced the resolution of these principles to enable programmed fate differentiation, distinguishing different stages of cell fate.
- We demonstrated the potential to detect RNA pathway alterations in the e-cigarette dataset and the ability to classify functional cell interactions in the rat myocardial infarction (MI) model dataset.
𝑻𝒆𝒓𝒎𝒊𝒏𝒐𝒍𝒐𝒈𝒚 𝑬𝒙𝒑𝒍𝒂𝒏𝒂𝒕𝒊𝒐𝒏:
• Subtype Classification: A method of categorizing cell types. Specific mature cells can differentiate into different subtypes under various conditions. For instance, CD4⁺ T cells can transition into Th1 cells, Th2 cells, Treg cells, or Tfh cells. These all belong to the broader category of T cells but represent distinct subcategories. For clarity, we define that if a cell type does not generate other subtypes, it is considered to have only one subtype.
• Lineage Tracing: A functional classification method for cells. It predicts the types of cells that a progenitor cell with differentiation potential will develop into. For example, hematopoietic cells may differentiate into monocytes or neutrophils.
• Programmed Fate Differentiation: A classification of cell fate stages. Within the same cell subtype or type, specific programmed events are initiated for a particular purpose, such as healthy cells undergoing apoptosis or ferroptosis. This is a new topic we trying to talk for single cell.
Discover of Biology:
• We mapped the trajectory of neuronal programmed cell death and identified neuron populations primarily undergoing ferroptosis.
• Key ferroptosis-driving genes, including Ctsb, Mtdh, Ndrg1, Smad7, and others, were highlighted as potential therapeutic targets. By characterizing these pathways, we provide new insights into the regulation of ferroptosis, which is essential for rescuing neurons and mitigating damage in affected brain areas.
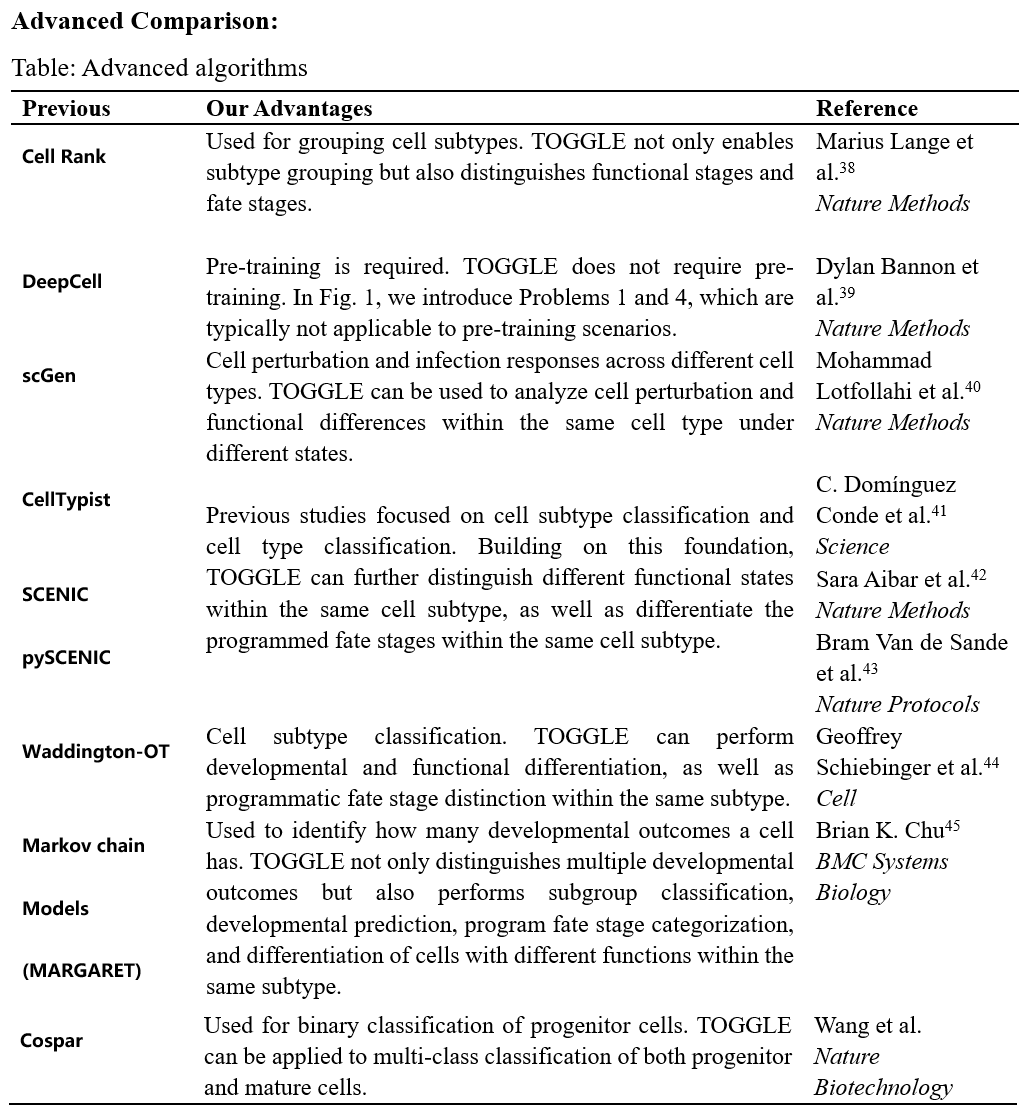
𝑨𝒅𝒗𝒂𝒏𝒄𝒆𝒅 𝑪𝒐𝒎𝒑𝒂𝒓𝒊𝒔𝒐𝒏:
5.Raw data
We provide a format-converted dataset on the cloud drive.
If you wish to access the original dataset, you can do so through the following link.
𝙏𝙝𝙖𝙣𝙠𝙨 𝙩𝙤 𝙩𝙝𝙚𝙨𝙚 𝙖𝙪𝙩𝙝𝙤𝙧𝙨 𝙛𝙤𝙧 𝙩𝙝𝙚𝙞𝙧 𝙨𝙥𝙞𝙧𝙞𝙩 𝙤𝙛 𝙨𝙝𝙖𝙧𝙞𝙣𝙜. 𝙏𝙝𝙚𝙮 𝙖𝙧𝙚 𝙜𝙧𝙚𝙖𝙩 𝙥𝙚𝙤𝙥𝙡𝙚.
Hematopoietic Development (GSE140802)
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140802
Fibroblast Reprogramming (GSE99915)
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99915
E-Cigarette and Nicotine Exposures (GSE183614)
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183614
Rat Myocardial Infarction (MI) model (GSE253578)
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253578
Programmed Cell Death Trajectories Prediction in Neurons (GSE232429)
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232429
