Mature Functions: RNA Pathways Changes of Ecigarettes
1.Main: Adult mice-e-cigarette-only e-cigarette and air-only difference in genes
1.1.Calculate the correlation of RNA. (Python)
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "成年小鼠-只电子烟和空气.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=20,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'Adult mice - e-cigarette and air only.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'Adult mice - e-cigarette and air only.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
1.2.Cluster RNA using unsupervised methods. (Matlab)
%% Load data and Split to compute
MM0 = load('./result/Adult mice - e-cigarette and air only.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/Adult mice - e-cigarette and air only.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,10,2,2);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.03]
%filename = fullfile('result', 'Adult mice - e-cigarette and air only.csv');
writetable(count_result, filename);
1.3.Obtain group segmentation results. (Matlab)
%% 临近法激活
figure(4)
corr_matrix = relevance_generate(0.0140,4,cluster_map_matrix);
hi = heatmap(corr_matrix);
%% 编码
encode_result = encoder_corr_matrix(0.0141,0.0139,10,2,cluster_map_matrix);
figure(2)
hj = heatmap(encode_result);
%% 解码
figure(3)
[weighting_decode,decode_result] = decoder_corr_matrix(encode_result);
weighting_result = weighting_decode + decode_result;
hk = heatmap(decode_result);
hk.ColorLimits = [9,11]
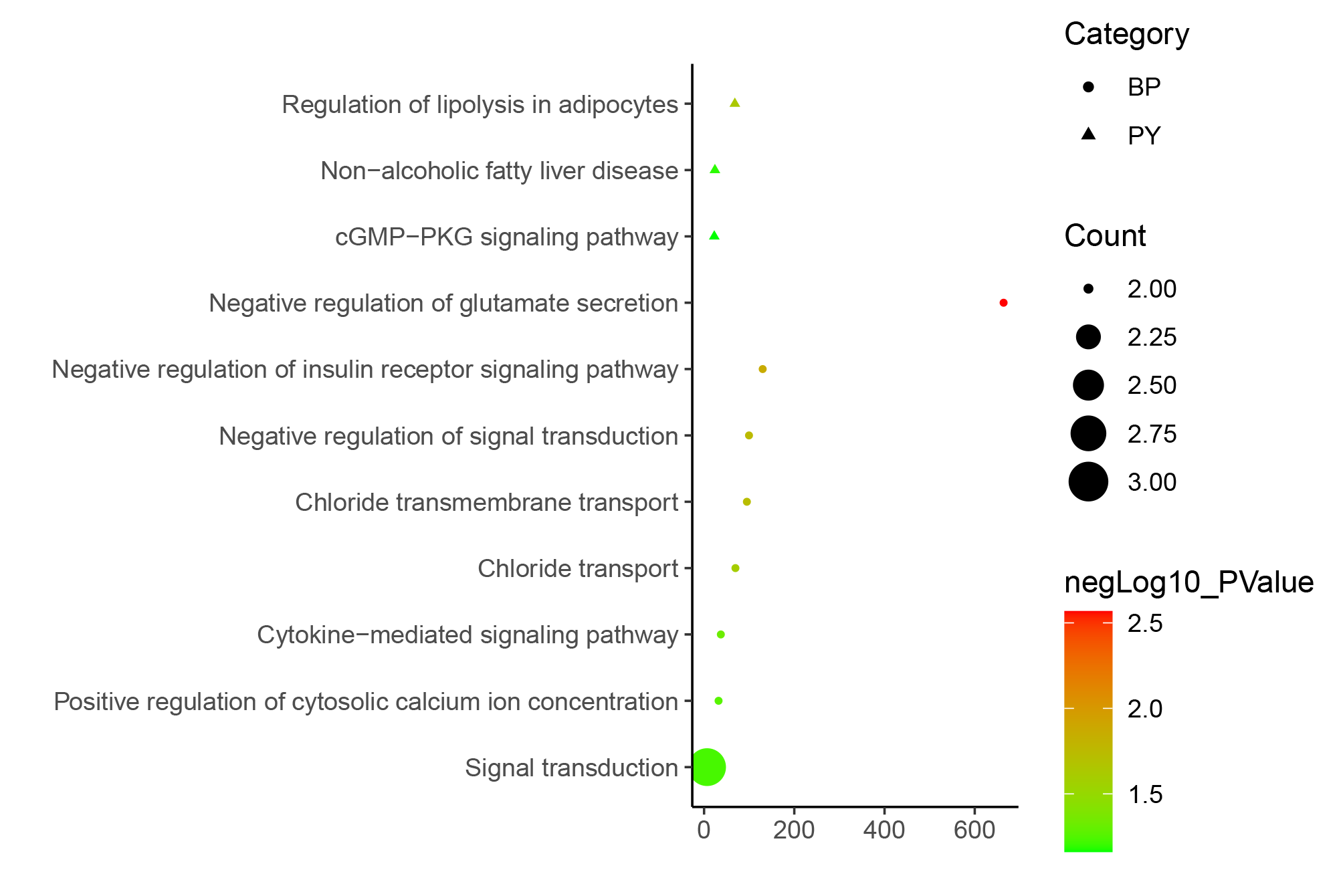
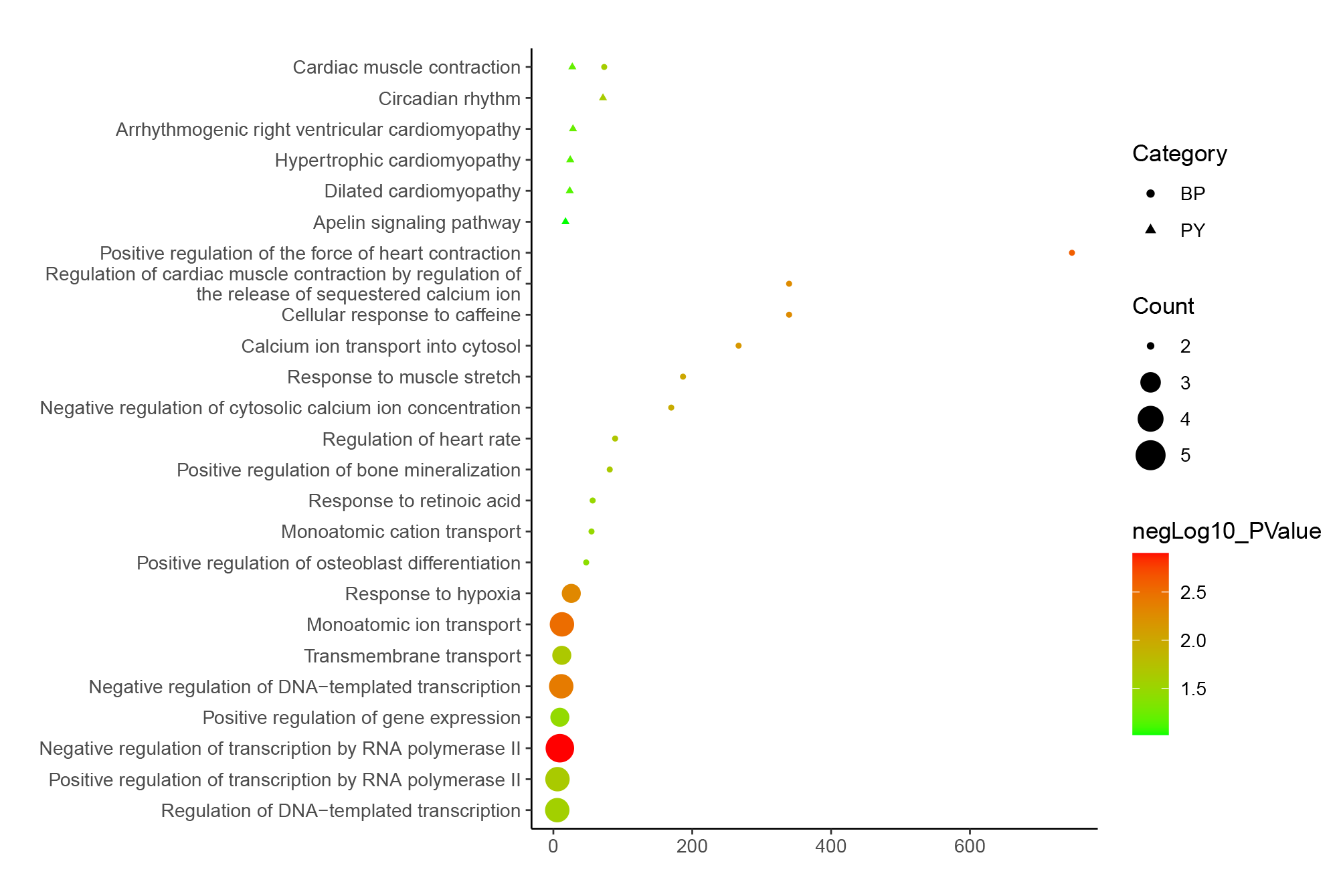
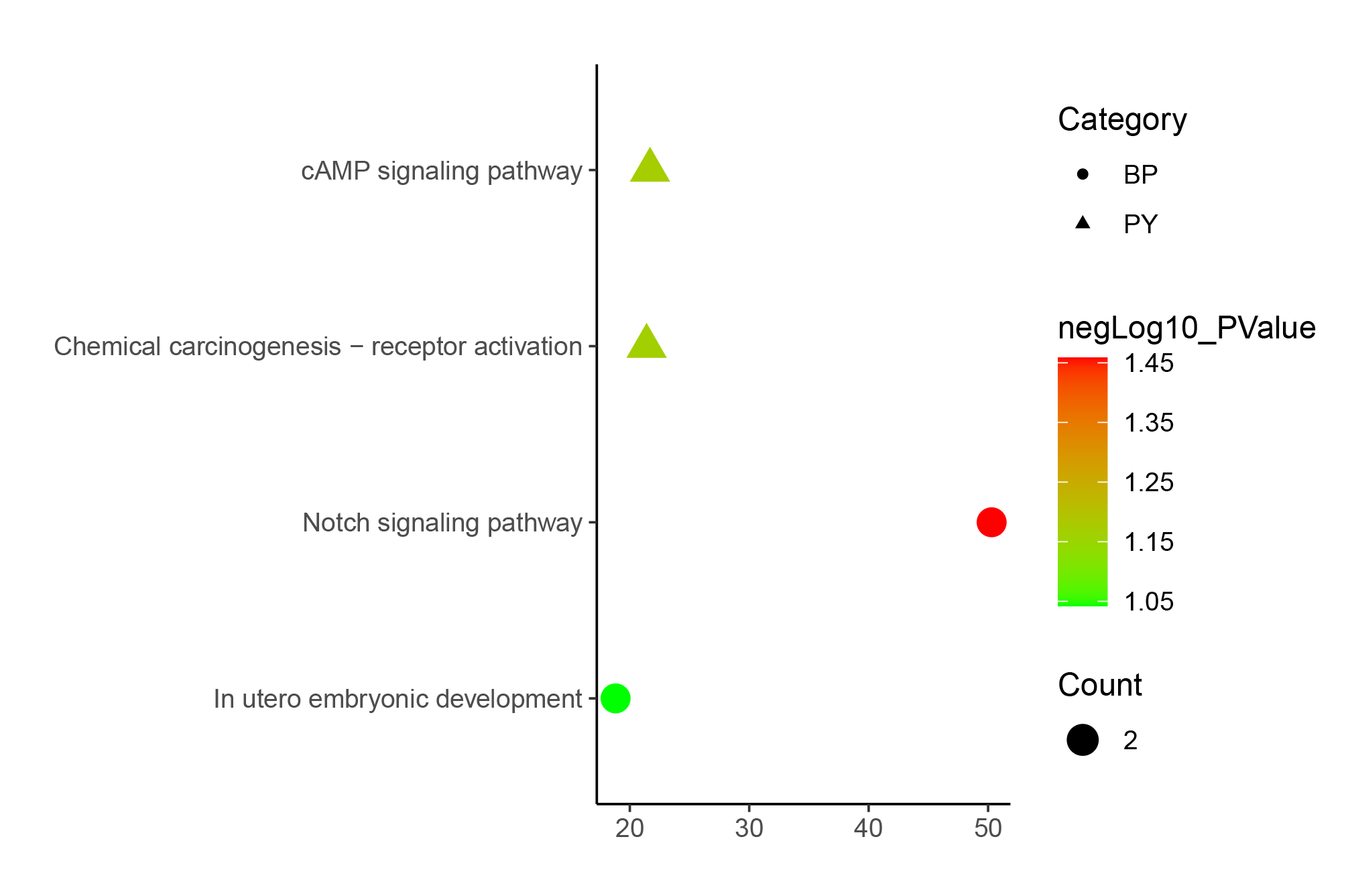
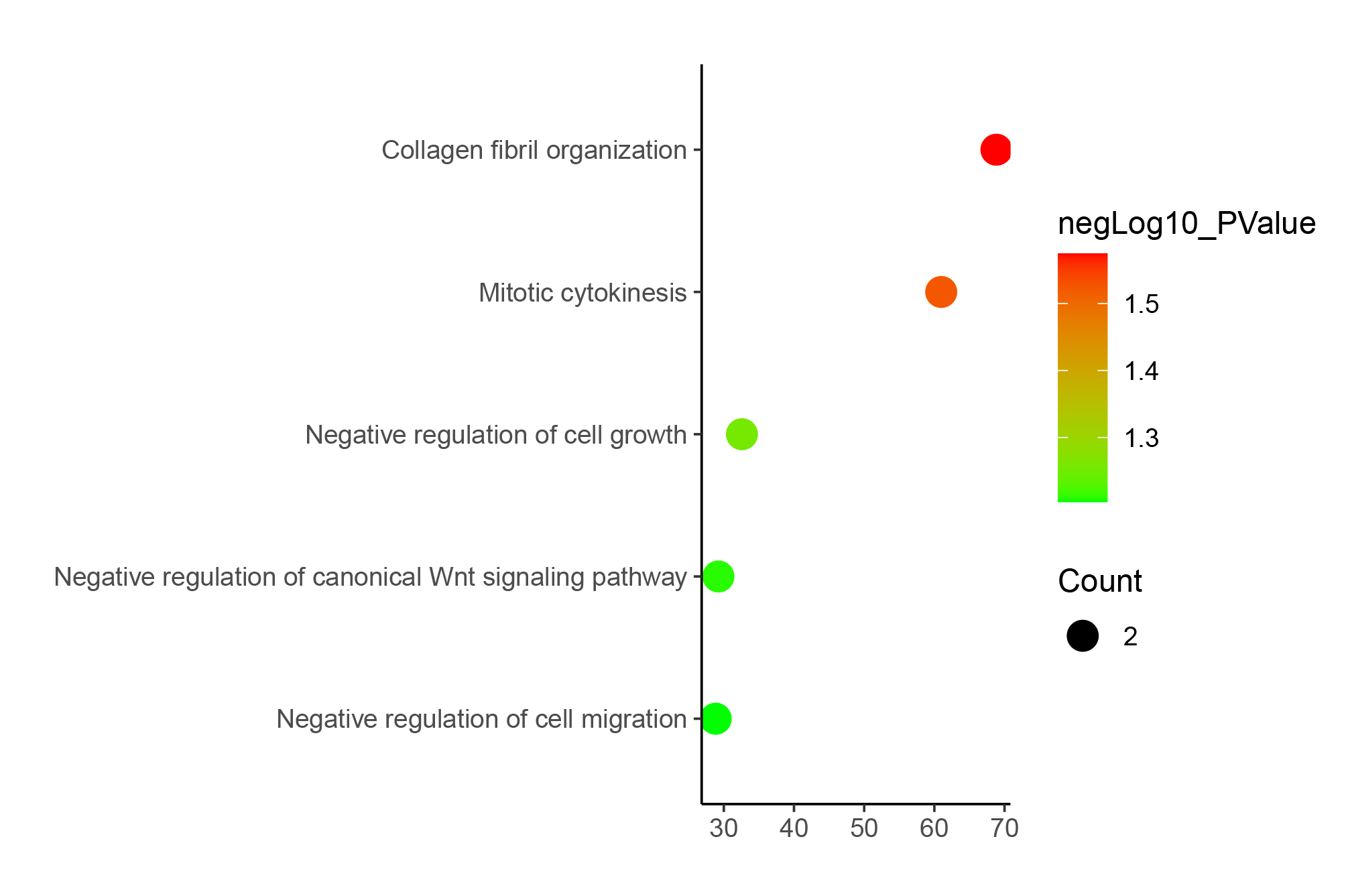
We selected a set shown in the article for testing, but there are actually many other ways to use it. Here are some examples that were not included in the article due to space limitations.
2.Other potential experiments.
Adult mice-Electronic cigarette (Python)
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "成年小鼠-电子烟.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(cigarette_data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=20,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import matplotlib
matplotlib.use('TkAgg') # 或 'Qt5Agg' 等
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib
# 设置后端
matplotlib.use('TkAgg')
# 绘制热图
sns.heatmap(cigarette_matrix_dense, cmap='Blues', vmin=0, vmax=0.00001)
plt.title('Distance Matrix')
plt.xlabel('Samples')
plt.ylabel('Samples')
plt.show()
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'Adult_mice-Electronic_cigarette.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'Adult_mice-Electronic_cigarette.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
%% Load data and Split to compute
MM0 = load('./result/Adult_mice-Electronic_cigarette.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/Adult_mice-Electronic_cigarette.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,25,5,5);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.00007]
filename = fullfile('result', 'Adult_mice-Electronic_cigarette_result.csv');
%writetable(count_result, filename);
Adult_mice-Electronic_freshair (Python)
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "成年小鼠-新鲜空气.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(cigarette_data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=20,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'Adult_mice-Electronic_freshair.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'Adult_mice-Electronic_freshair.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
(Matlab)
%% Load data and Split to compute
%% Load data and Split to compute
MM0 = load('./result/Adult_mice-Electronic_freshair.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/Adult_mice-Electronic_freshair.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,25,5,5);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.00007]
filename = fullfile('result', 'Adult_mice-Electronic_freshair_result.csv');
%writetable(count_result, filename);
Fresh air in children vs adults - Child mice - E-cigarettes
Child mice - aVc_c (Python)
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "幼年vs成年的新鲜空气-幼年小鼠-电子烟.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=2000,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'Child mice - aVc_c.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'Child mice - aVc_c.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
(Matlab)
%% Load data and Split to compute
MM0 = load('./result/Child mice - aVc_c.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/Child mice - aVc_c.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,10,2,2);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.002]
filename = fullfile('result', 'Child mice - aVc_c.csv');
%writetable(count_result, filename);
%% 临近法激活
figure(4)
corr_matrix = relevance_generate(0.00035,2,cluster_map_matrix);
corr_matrix = relevance_generate(0.0004,2,cluster_map_matrix);
hi = heatmap(corr_matrix);
%% 编码
encode_result = encoder_corr_matrix(0.00091,0.00080,10,2,cluster_map_matrix);
figure(2)
hj = heatmap(encode_result);
%% 解码
figure(3)
[weighting_decode,decode_result] = decoder_corr_matrix(encode_result);
weighting_result = decode_result;
hk = heatmap(decode_result);
hk.ColorLimits = [23,25]
Child vs adult electronic cigarettes - Child mice - electronic cigarettes
Child mice - c_c (Python) —
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "幼年vs成年的电子烟-幼年小鼠-电子烟.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=2000,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'Child mice - c_c.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'Child mice - c_c.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
(Matlab)
%% Load data and Split to compute
MM0 = load('./result/Child mice - c_c.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/Child mice - c_c.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,25,5,5);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.0003]
filename = fullfile('result', 'Child mice - c_c.csv');
%writetable(count_result, filename);
%% 临近法激活
figure(4)
corr_matrix = relevance_generate(0.00035,2,cluster_map_matrix);
corr_matrix = relevance_generate(0.0004,2,cluster_map_matrix);
hi = heatmap(corr_matrix);
%% 编码
encode_result = encoder_corr_matrix(0.00051,0.00050,10,2,cluster_map_matrix);
figure(2)
hj = heatmap(encode_result);
%% 解码
figure(3)
[weighting_decode,decode_result] = decoder_corr_matrix(encode_result);
weighting_result = decode_result;
hk = heatmap(decode_result);
hk.ColorLimits = [35,37]
Electronic cigarettes vs. fresh air - adult mice - electronic cigarettes
aVc_c (Python) —
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
#初始化函数,将kailin转至工作目录。如果此前初始化过,那么在再次运行def kl_initialize(0)时,
#则拒绝初始化,避免套娃。运行def kl_initialize(1)时,强制重新初始化。
kl.kl_initialize(0)
#获取kailin工作的根目录
parent_directory_origin = kl.kl_settings.parent_directory_origin
print(parent_directory_origin)
#改进:
#添加一个cluster模式
#选择进行Lineage Tracing还是Cluster,并给出可用的列表
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Cluster',1)
print(current_folder)
#选择要使用哪个样本
choosen_sample = "e-cigarettes_difference expression"
#选择.h5ad文件
h5ad_filename = "电子烟vs新鲜空气-幼年小鼠-电子烟.xlsx"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
current_folder_input = os.path.join(current_folder_input, 'database')
current_folder_input = os.path.join(current_folder_input, 'Clustering_sample')
current_folder_input = os.path.join(current_folder_input, 'e-cigarettes_difference_expression')
current_save_path = os.path.join(current_folder_input, 'result')
current_folder_input = os.path.join(current_folder_input, 'data')
current_folder_input = os.path.join(current_folder_input, h5ad_filename)
print(current_folder_input)
import pandas as pd
import anndata as ad
data = pd.read_excel(current_folder_input)
print(data)
# Selecting all rows and all columns after the first in the original data
cigarette_data_X = data.iloc[:, 1:]
# Displaying the first few rows to confirm the selection
print(cigarette_data_X)
cigarette_adata = ad.AnnData(X=cigarette_data_X.values)
print(cigarette_adata)
from kailin.omic_functions import matrix
cigarette_matrix = matrix.distance_matrix(
cigarette_adata,
round_of_smooth=1,
neighbor_N=2000,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
import scipy.sparse
# 假设 cigarette_matrix 是一个稀疏矩阵
cigarette_matrix_dense = cigarette_matrix.toarray() # 转换为非稀疏 numpy 数组
print(cigarette_matrix_dense)
import scipy.io as sio
current_save_path_mat = os.path.join(current_save_path, 'aVc_c.mat')
sio.savemat(current_save_path_mat, {'cigarette_matrix_dense': cigarette_matrix_dense})
label = data.iloc[:, 0]
print(label)
import os
import pandas as pd
# 提取 data 的第一列
label = data.iloc[:, 0] # 如果 data 是 numpy 数组
# 如果 data 是 pandas DataFrame,则用 label = data.iloc[:, 0]
# 确保保存路径存在
save_folder = 'current_save_path'
if not os.path.exists(current_save_path):
os.makedirs(current_save_path)
# 设置保存文件的完整路径
current_save_path_csv = os.path.join(current_save_path, 'aVc_c.csv')
# 保存为 CSV 文件
pd.DataFrame(label).to_csv(current_save_path_csv, index=False)
print(f"文件已保存到 {current_save_path_csv}")
(Matlab)
%% Load data and Split to compute
MM0 = load('./result/aVc_c.mat');
MM0 = MM0.cigarette_matrix_dense;
%% 读取要排序的对象
count_=readtable('./result/aVc_c.csv');
%% 得到边界划分点
[p,splitlist] = binary_corr_sorting(MM0,20,25,5,5);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0, 0.0003]
filename = fullfile('result', 'aVc_c.csv');
%writetable(count_result, filename);
%% 临近法激活
figure(4)
corr_matrix = relevance_generate(0.0005,2,cluster_map_matrix);
hi = heatmap(corr_matrix);
%% 编码
encode_result = encoder_corr_matrix(0.0005,0.00049,10,1,cluster_map_matrix);
figure(2)
hj = heatmap(encode_result);
%% 解码
figure(3)
[weighting_decode,decode_result] = decoder_corr_matrix(encode_result);
weighting_result = decode_result;
hk = heatmap(decode_result);
hk.ColorLimits = [36,38]
