Mature Functions: RNA Map of Myocardial
The default data file storage directory is C:/GEOANALYSIS/GSE253768
Highly recommended to use Rstudio. Need to select the environment which containing anndata, in Tools > Global options > Python > Python interpreter
1.Preprocessing and generate h5ad file.
1.1.Load required packages (R)
library(Seurat)
library(multtest)
library(dplyr)
library(ggplot2)
library(patchwork)
library(tidyverse)
library(future)
library(harmony)
library(RColorBrewer)
# Create a vector to read files
setwd("C:/GEOANALYSIS/GSE253768")
## Save the file names in the folder to 'dir_name'
dir_name <- list.files(pattern = "\\.csv$") # Match only CSV files
## View 'dir_name'
dir_name
#[1] "MI1.csv" "MI2.csv" "Sham1.csv" "Sham2.csv"
## Assign names to 'dir_name' (use file name without extension)
names(dir_name) <- gsub("\\.csv$", "", dir_name)
## View the renamed 'dir_name'
dir_name
## MI1 MI2 Sham1 Sham2
##"MI1.csv" "MI2.csv" "Sham1.csv" "Sham2.csv"
## Batch data processing
scRNAlist <- list()
for (i in 1:length(dir_name)) {
# Read CSV file
counts <- read.csv(file = dir_name[i], row.names = 1) # Use the first column as row names
counts <- as.matrix(counts) # Convert to matrix format
# Create Seurat object and add file name as a label
scRNAlist[[i]] <- CreateSeuratObject(
counts = counts,
min.cells = 3,
min.features = 300,
project = names(dir_name)[i]
)
}
# Check the Seurat object list
scRNAlist
# Calculate mitochondrial and red blood cell proportions in batch
for(i in 1:length(scRNAlist)){
sc <- scRNAlist[[i]]
# Calculate mitochondrial proportion
sc[["mt_percent"]] <- PercentageFeatureSet(sc, pattern = "^Mt-")
# Calculate red blood cell proportion
HB_genes <- c("Hba1","Hba2","Hbb","Hbd","Hbe1","Hbg1","Hbg2","Hbm","Hbq1","Hbz")
HB_m <- match(HB_genes, rownames(sc@assays$RNA))
HB_genes <- rownames(sc@assays$RNA)[HB_m]
HB_genes <- HB_genes[!is.na(HB_genes)]
sc[["HB_percent"]] <- PercentageFeatureSet(sc, features=HB_genes)
# Assign 'sc' back to scRNAlist[[i]]
scRNAlist[[i]] <- sc
# Remove 'sc'
rm(sc)
}
1.2.Perform a simple merge and then plot quality control (R)
CI <- merge(scRNAlist[[1]], y=c(scRNAlist[[2]], scRNAlist[[3]], scRNAlist[[4]]))
head(colnames(CI))
unique(sapply(X = strsplit(colnames(CI), split = "_"), FUN = "[", 1))
plot1 <- FeatureScatter(CI, feature1 = "nFeature_RNA", feature2 = "mt_percent")
plot2 <- FeatureScatter(CI, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
CombinePlots(plots = list(plot1, plot2))# QC plot 1
VlnPlot(CI, features = c("mt_percent", "nFeature_RNA", "nCount_RNA", "HB_percent"), ncol = 4, pt.size=0)# QC plot 2
VlnPlot(CI, features = c("mt_percent", "nFeature_RNA", "nCount_RNA", "HB_percent"), ncol = 4, pt.size=0.5)# QC plot 3
# Filter cells in batch
scRNAlist <- lapply(X = scRNAlist, FUN = function(x){
x <- subset(x,
subset = nFeature_RNA > 200 & nFeature_RNA < 5000 &
mt_percent < 10 &
HB_percent < 5 &
nCount_RNA < quantile(nCount_RNA,0.97))})
# Merge Seurat objects
scRNAlist <- merge(scRNAlist[[1]], y=c(scRNAlist[[2]], scRNAlist[[3]], scRNAlist[[4]]))
# Select highly variable genes and perform dimensionality reduction
scRNAlist <- NormalizeData(scRNAlist) %>%
FindVariableFeatures(selection.method = "vst",nfeatures = 3000) %>%
ScaleData() %>%
RunPCA(npcs = 30, verbose = T)
# Integrate using Harmony
testAB.integrated <- RunHarmony(scRNAlist, group.by.vars = "orig.ident")
# Copy 'orig.ident' to 'Sample'
testAB.integrated@meta.data$Sample <- testAB.integrated@meta.data$orig.ident
# Copy 'orig.ident' to 'Group' and remove numbers
testAB.integrated@meta.data$Group <- gsub("[0-9]", "", testAB.integrated@meta.data$orig.ident)
# Check the updated metadata
head(testAB.integrated@meta.data)
# Add grouping information after integration
metadata <- testAB.integrated@meta.data
write.csv(metadata, file="meta.data.csv")# Export and save
#testAB.integrated@meta.data <- metadata
# Perform clustering
testAB.integrated <- FindNeighbors(testAB.integrated, reduction = "harmony", dims = 1:15) %>% FindClusters(resolution = 0.18)#15群
# Perform UMAP/tSNE dimensionality reduction
testAB.integrated <- RunTSNE(testAB.integrated, reduction = "harmony", dims = 1:15)
testAB.integrated <- RunUMAP(testAB.integrated, reduction = "harmony", dims = 1:25)
# Save
save(testAB.integrated, metadata, file = "MI Cell-15 clusters.Rdata")
# Export markers
testAB.integrated <- JoinLayers(testAB.integrated)
CI.markers <- FindAllMarkers(testAB.integrated, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25)
write.csv(CI.markers, file="MI Cell marker.csv")
# Naming the 15 clusters
new.cluster.ids <- c("Fibroblasts", "Cardiomyocytes", "Endothelial cells",
"Cardiomyocytes", "Smooth muscle cells", "Macrophages",
"T cells", "Endothelial cells", "Endothelial cells",
"Mesothelial cells", "Macrophages", "Endothelial cells",
"Schwann cells", "Myofibroblast", "Endothelial cells")
names(new.cluster.ids) <- levels(testAB.integrated)
testAB.integrated <- RenameIdents(testAB.integrated, new.cluster.ids)
testAB.integrated$clusters2 <- testAB.integrated@active.ident
save(testAB.integrated, metadata, file = "MI Cell-15 clusters.Rdata")
# Export the count of each cluster
Table1 <- table(testAB.integrated$Group, testAB.integrated$clusters2)
Table2 <- table(testAB.integrated$Sample, testAB.integrated$clusters2)
write.table(Table1, file = "Cell counts in each group.txt", sep ="\t")
write.table(Table2, file = "Cell counts in each sample.txt", sep ="\t")
# Export UMAP images from preliminary analysis
cell_type_cols <- c("#B383B9", "#F5CFE4","#EE934E","#F5D2A8","#fced82","#D2EBC8","#7DBFA7","#AECDE1","#3c77af")
p1 <- DimPlot(testAB.integrated, reduction = "umap", group.by = "clusters2", pt.size=0.5, label = T,repel = TRUE, raster=FALSE, cols = cell_type_cols) + labs(x = "UMAP1", y = "UMAP2") + theme(panel.border = element_rect(fill=NA,color="black", size=1, linetype="solid"), axis.text.y = element_blank(), axis.ticks.y = element_blank(), axis.text.x = element_blank(), axis.ticks.x = element_blank())
ggsave(filename = "Preliminary grouping of MI - overall.pdf", plot = p1, device = 'pdf', width = 21, height = 18, units = 'cm')
testAB.integrated <- subset(testAB.integrated,idents=c("Fibroblasts","Myofibroblast"),invert = FALSE)
testAB.integrated$RNA_snn_res.0.18 <- NULL
testAB.integrated$clusters1 <- NULL
testAB.integrated$clusters2 <- NULL
testAB.integrated$seurat_clusters <- NULL
#Re-finding highly variable genes
testAB.integrated <- NormalizeData(testAB.integrated) %>%
FindVariableFeatures(selection.method = "vst",nfeatures = 3000) %>%
ScaleData() %>%
RunPCA(npcs = 30, verbose = T)
# Save
save(testAB.integrated, file = "MI-FibroblastCell.Rdata")
1.3.Exporting RNA files (R)
# Load data
load("C:/GEOANALYSIS/GSE253768/MI-FibroblastCell.Rdata")
# Downgrade the matrix to an older version if necessary
testAB.integrated[["RNA"]] <- as(object = testAB.integrated[["RNA"]], Class = "Assay")
# Extract fibroblasts from the myocardial infarction group
testAB.integrated_MI <- subset(testAB.integrated,idents=c("Fibroblasts"),invert = FALSE)
Idents(testAB.integrated) <- "Group"
testAB.integrated_MI <- subset(testAB.integrated,idents=c("MI"),invert = FALSE)
# Recalculate highly variable genes
testAB.integrated_MI <- NormalizeData(testAB.integrated_MI) %>%
FindVariableFeatures(selection.method = "vst",nfeatures = 3000) %>%
ScaleData() %>%
RunPCA(npcs = 30, verbose = T)
# Extract the expression matrix of highly variable genes
# Extract the expression matrix of highly variable genes from the data slot
hvg_genes <- VariableFeatures(testAB.integrated_MI) # Get the names of highly variable genes
hvg_matrix <- testAB.integrated_MI@assays$RNA@data[hvg_genes, ] # Extract the expression data of these highly variable genes
# Transpose the matrix
hvg_matrix_transposed <- t(hvg_matrix)
# Create a new Seurat object and save the transposed matrix into it
testAB.integrated_MI <- CreateSeuratObject(counts = hvg_matrix_transposed)
# Select highly variable genes and perform dimensionality reduction
testAB.integrated_MI <- NormalizeData(testAB.integrated_MI) %>%
FindVariableFeatures(selection.method = "vst",nfeatures = 3000) %>%
ScaleData() %>%
RunPCA(npcs = 30, verbose = T)
# Perform UMAP dimensionality reduction
# Smaller n.neighbors: tighter clusters; larger n.neighbors: more uniform distribution
# Smaller min.dist: tighter clusters; larger min.dist: more uniform distribution
# Smaller spread: more concentrated plot; larger spread: better separation between clusters
testAB.integrated_MI <- RunUMAP(testAB.integrated_MI, dims = 1:15,
spread = 2, n.neighbors = 100, min.dist = 0.8)# This separates gene groups into 3 distinct clusters
UMAPPlot(testAB.integrated_MI,label=T)
# Save
save(testAB.integrated_MI, file = "FibroblastCellMatrix Transposition of MI.Rdata")
### Ensure the matrix includes all cells
DefaultAssay(testAB.integrated_MI) <- "RNA"
testAB.integrated_MI[["RNA"]] <- as(object = testAB.integrated_MI[["RNA"]], Class = "Assay")# Convert to version 4 matrix
sceasy::convertFormat(
testAB.integrated_MI,
from = "seurat",
to = "anndata",
outFile = "MI_fiberRNA.h5ad"
)
Save the R variable table as variables.Rdata.
2.Calculate functional mapping. (Python)
Move C:/GEOANALYSIS/GSE253768/MI_fiberRNA.h5ad to [Path of LittleSnowFox]/database/Clustering_sample/fibroblasts/data/
import numpy as np
import os
import LittleSnowFox as kl
#import matlab.engine
#eng = matlab.engine.start_matlab()
print(kl.__version__)
kl.kl_initialize(0)
parent_directory_origin = kl.kl_settings.parent_directory_origin
current_folder = kl.workcatalogue.choosemode_kl(parent_directory_origin,'Clustering',1)
#选择要使用哪个样本
choosen_sample = "fibroblasts"
#选择.h5ad文件
h5ad_filename = "MI_fiberRNA.h5ad"
#运行自带的示例,并获取稀疏矩阵
#这里需要做非示例的函数进去
current_folder_input = current_folder
orig_adata,loading_directory,distance_matrix = kl.preprocessing.kl_dense_matrix_sample(
choosen_sample,
h5ad_filename,
"draw",
current_folder_input,
round_of_smooth=1,
neighbor_N=13000,
beta=0.1,
truncation_threshold=0.001,
save_subset=True,
use_existing_KNN_graph=False,
compute_new_Smatrix=True,
use_full_Smatrix = True,
)
#orig_adata,loading_directory,distance_matrix_sparse = kl.preprocessing.kl_dense_matrix_sample(choosen_sample,h5ad_filename,"draw",current_folder_input)
import os
import scipy.io
current_folder_result = os.path.join(loading_directory, 'result')
print(current_folder_result)
mat_name = "distance_matrix_RNA.mat"
mat_path = os.path.join(current_folder_result, mat_name)
scipy.io.savemat(mat_path, {"distance_matrix": distance_matrix})
import pandas as pd
obs_names_list = orig_adata.obs_names.tolist()
var_names_list = orig_adata.var_names.tolist()
index = orig_adata.obs.index
df_orig_adata = pd.DataFrame({
"Index": index,
})
print(df_orig_adata)
csv_name = "orig_ident_RNA.csv"
csv_path = os.path.join(current_folder_result, csv_name)
print(csv_path)
df_orig_adata.to_csv(csv_path, index=False)
3.Unsupervised learning (Matlab) —
%% Load data and Split to compute
%% Load data and Split to compute
MM0 = load('./result/distance_matrix_RNA.mat');
MM0 = MM0.distance_matrix;
%% 读取要排序的对象
count_=readtable('./result/orig_ident_RNA.csv');
%% 得到边界划分点
%[p,splitlist] = binary_corr_sorting(MM0,20,125,5,5);
[p,splitlist] = binary_corr_sorting(MM0,20,50,5,5);
%% 对划分点去重
[uniqueList, ~, ~] = unique(splitlist, 'stable');
%% 对相似度矩阵排序
MM=MM0(p,p);
split=[];
%% 重排count_result
count_result=count_(p,:);
split_simple=uniqueList;
%% 第一个起始位点置为1
split_simple(1)=1;
split_simple=[split_simple,length(MM0)];
%% 计算均值矩阵
[simple_matrix]=sample_computing(count_result,split_simple,MM,"mean");
%% 合并成小矩阵
ClusterReslut=cluster_map(split_simple,simple_matrix,0,0.0002,0);
count_result.Result = ClusterReslut;
%重排小矩阵
[cluster_map_matrix] = genetic_encoder( ...
simple_matrix, ...
60, ...% nPop = 50; % 种群规模大小为30
1, ...% nPc = 1; % 子代规模的比例0.8
200, ...% maxIt = 200; % 最大迭代次数
5 ...% cycletimes = 200; % 循环计算次数
);
%重拍小矩阵方案2
% 创建行和列标签(示例)
%row_labels = cluster_map_label;
%column_labels = cluster_map_label;
% 使用 heatmap 函数并传递相应参数
h = heatmap(cluster_map_matrix);
%h.YDisplayLabels = row_labels; % 设置行标签
%h.XDisplayLabels = column_labels; % 设置列标签
h.ColorLimits = [0,0.001]%
%% 临近法激活
corr_matrix = relevance_generate(0.0001,1,cluster_map_matrix);
hi = heatmap(corr_matrix);
%% 编码
% encode_result = encoder_corr_matrix(0.0011,0.001,10,10,cluster_map_matrix);
encode_result = encoder_corr_matrix(0.00011,0.00010,10,1,cluster_map_matrix);
figure(2)
hj = heatmap(encode_result);
%% 解码
figure(3)
[weighting_decode,decode_result] = decoder_corr_matrix(encode_result);
weighting_result = decode_result + 2*weighting_decode;
hk = heatmap(weighting_result);
hk.ColorLimits = [60,65]
writetable(count_result, './result/result_RNA.csv');
4.Generate RNA map (R)
Read the R variable table as variables.Rdata.
Move result_RNA.csv from [Little Snow Fox installation directory]\database\Clustering_sample\fibroblasts\result to C:\GEOANALYSIS\GSE253768.
# Import the results
index_result <- read.csv("result_RNA.csv")
## Ensure the 'Index' in the table matches the cell names in the Seurat object
## Set 'Index' as row names for easier operations
rownames(index_result) <- index_result$Index
## Map the 'Result' column to the metadata of the Seurat object using 'Index'
metadata <- testAB.integrated_MI@meta.data # Get the metadata of the Seurat object
metadata$Result <- index_result[rownames(metadata), "Result"]
## Update the metadata of the Seurat object
testAB.integrated_MI@meta.data <- metadata
## Check the updated metadata
head(testAB.integrated_MI@meta.data)
# Get the metadata of the Seurat object
metadata <- testAB.integrated_MI@meta.data
# Create a 'Fenqun' column and group based on 'Result' column values
metadata$Fenqun <- NA # Initialize the 'Fenqun' column
# Use conditional statements to group 'Result' column values by ranges
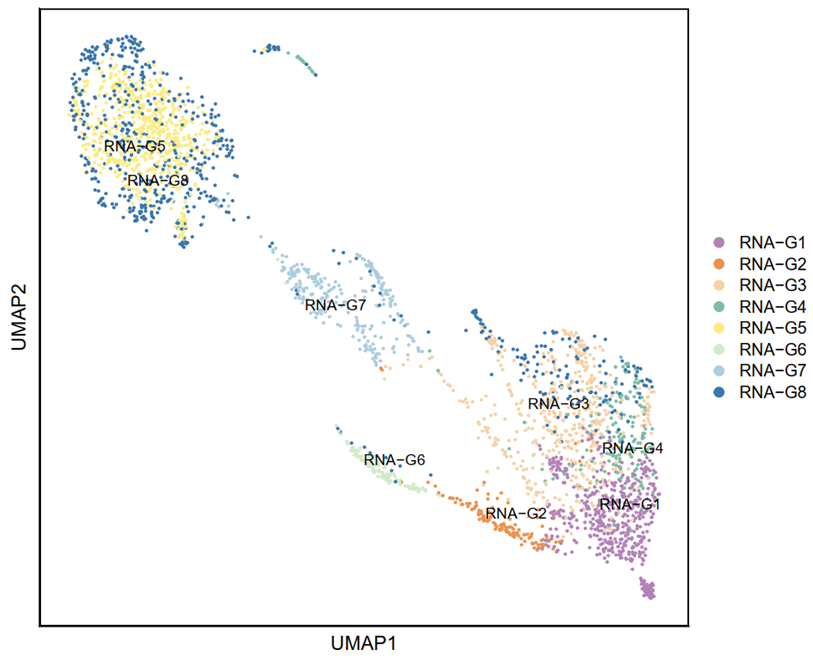
metadata$Fenqun[metadata$Result >= 1 & metadata$Result <= 16] <- "RNA-G1"
metadata$Fenqun[metadata$Result >= 17 & metadata$Result <= 20] <- "RNA-G2"
metadata$Fenqun[metadata$Result >= 21 & metadata$Result <= 35] <- "RNA-G3"
metadata$Fenqun[metadata$Result >= 36 & metadata$Result <= 41] <- "RNA-G4"
metadata$Fenqun[metadata$Result >= 42 & metadata$Result <= 61] <- "RNA-G5"
metadata$Fenqun[metadata$Result >= 62 & metadata$Result <= 65] <- "RNA-G6"
metadata$Fenqun[metadata$Result >= 66 & metadata$Result <= 72] <- "RNA-G7"
metadata$Fenqun[metadata$Result >= 73 & metadata$Result <= 92] <- "RNA-G8"
# Update the modified metadata into the Seurat object
testAB.integrated_MI@meta.data <- metadata
# View results
head(testAB.integrated_MI@meta.data$Fenqun)
# Plotting
cell_type_cols <- c("#B383B9", "#EE934E","#F5D2A8","#7DBFA7","#fced82","#D2EBC8","#AECDE1","#3c77af")
p1 <- DimPlot(testAB.integrated_MI, reduction = "umap", group.by = "Fenqun", pt.size=0.5, label = T,repel = TRUE, raster=FALSE, cols = cell_type_cols) + labs(x = "UMAP1", y = "UMAP2") + theme(panel.border = element_rect(fill=NA,color="black", size=1, linetype="solid"), axis.text.y = element_blank(), axis.ticks.y = element_blank(), axis.text.x = element_blank(), axis.ticks.x = element_blank())
ggsave(filename = "RNA clustering UMAP of MI.pdf", plot = p1, device = 'pdf', width = 21, height = 18, units = 'cm')
Save
metadata <- testAB.integrated_MI@meta.data
write.csv(metadata, file="Myocardial Fibroblast Cell RNA Clustering Results.csv")
